Abstract
Background: CPX-351 (Vyxeos) is a liposomal combination of daunorubicin and cytarabine that was FDA approved in 2017 for treatment of adults with newly diagnosed therapy-related acute myeloid leukemia (t-AML) or AML with myelodysplasia-related changes (AML-MRC). Genomic predictors of response to CPX-351 have not been described. TP53 mutations are uncommon in de novo AML, but relatively enriched in the t-AML and AML-MRC populations for which CPX-351 is approved (Christiansen DH et al. JCO 2001; Devillier R et al. Oncotarget 2015). As TP53 mutations confer resistance to conventional daunorubicin and cytarabine chemotherapy, we sought to determine whether TP53 mutations confer resistance to CPX-351.
Methods: This is a retrospective, multi-center review of patients who received at least 1 cycle of induction chemotherapy with CPX-351 at Memorial Sloan Kettering Cancer Center (MSKCC), Moffitt Cancer Center (MCC), and Weill Cornell Medical College (WCMC). 101 patients were identified at MSKCC (n=22), MCC (n=44), and WCMC (n=35). Responses to CPX-351 were graded using European Leukemia Net (ELN) response criteria. (Döhner H et al Blood 2017) Immunophenotypic minimal residual disease (MRD) was identified in bone marrow aspirates (BMA) by multiparameter flow cytometry in 43 patients (MSKCC n=22, WCMC n=21). Any level of residual disease was considered MRD+. Molecular analysis was obtained from pre-induction BMA by next-generation sequencing using 21, 32, 49, or 400 gene panels, all of which included TP53. Cytogenetics/FISH were performed using standard techniques. Fisher's exact tests were used to determine significance and are two-tailed. Kaplan-Meier (KM) analysis with log-rank test was performed to estimate overall survival (OS).
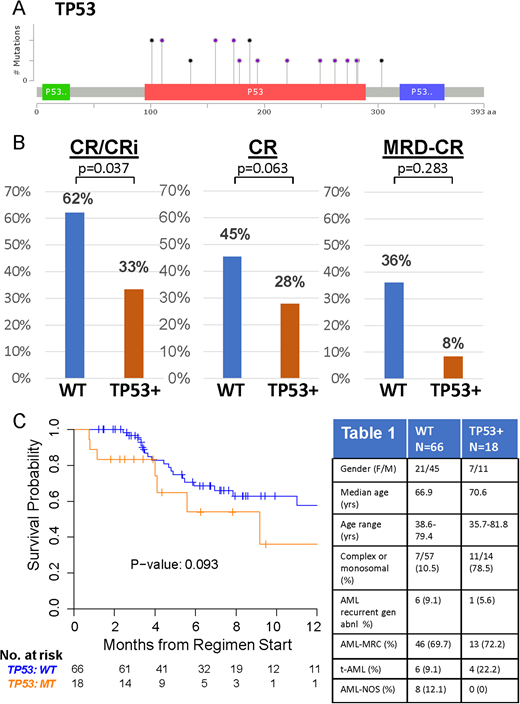
Results: Patient characteristics are in Table 1. 84/101 (83.1%) had baseline molecular profiling prior to CPX-351. TP53 mutations were identified in 18/84 (21.4% of patients). Analysis of additional co-mutations and responses will be presented by our collaborators (Talati et al., ASH 2018 submitted). TP53 mutations (19/20 distinct mutations) clustered in the DNA binding domain (Figure 1A). TP53 mutations are significantly associated with complex or monosomal karyotypes (p<0.001, see Table 1). Following 1-2 cycles of CPX-351 induction, complete responses (CR) combined with complete responses with incomplete blood count recovery (CRi) were significantly higher in TP53 wild-type (WT) patients compared to TP53 mutant patients. 62% WT (41/66) achieved CR/CRi, in comparison to 33% (6/18) TP53 mutant CR/CRi, p=0.0353, odds ratio=0.3049 (95 % CI:0.1016 to 0.9150). (Figure 1B). Responses for CR alone also trended in favor of TP53 WT patients. 45.5% (30/66) WT achieved CR, while 27.8% TP53 mutant (5/18) achieved CR, P=0.2806. Responses for MRD negative CR also favored WT patients, 36% MRD-CR WT (9/25), 8.3% MRD-CR TP53 mutant (1/12), although these results were not statistically significant likely due to low numbers of (n=37) who underwent molecular profiling and later MRD analysis. OS was not significantly different (Figure 3B, p=0.093), but also trended towards favoring WT over TP53 mutant patients. Additional data with longer-term follow-up will be presented.
Conclusion: Our data demonstrate that AML patients with TP53 mutations have lower rates of CR/CRi than WT patients after 1-2 cycles of CPX-351. While our study is retrospective and limited by small numbers, our data overall support the hypothesis that resistance to liposomal daunorubicin and cytarabine chemotherapy is common in AML patients with TP53 mutations. Future clinical studies are needed to determine optimal therapy for these patients.
Figure 1. A) Map of TP53 mutations in patients treated with CPX-351. Black circles represent truncating mutations; purple circles represent non-synonymous single nucleotide variants or splice site mutations. B) Rates of CR/CRi (left), CR (middle), and MRD negative CR in WT (n=66) and TP53 mutant (n=18) patients treated with CPX-351. C) OS for WT and TP53 mutant patients treated with CPX-351. Table 1. Patient Characteristics.
Goldberg:AROG: Research Funding; Celgene: Research Funding; Abbvie: Research Funding; Pfizer: Research Funding. Desai:Argenx: Consultancy; Cellerant Inc: Consultancy. Sallman:Celgene: Research Funding, Speakers Bureau. Roboz:Novartis: Consultancy; Celltrion: Consultancy; Novartis: Consultancy; Orsenix: Consultancy; Eisai: Consultancy; Sandoz: Consultancy; Pfizer: Consultancy; Roche/Genentech: Consultancy; AbbVie: Consultancy; Celgene Corporation: Consultancy; Aphivena Therapeutics: Consultancy; Orsenix: Consultancy; Bayer: Consultancy; Celltrion: Consultancy; Daiichi Sankyo: Consultancy; Bayer: Consultancy; Argenx: Consultancy; Celgene Corporation: Consultancy; Otsuka: Consultancy; AbbVie: Consultancy; Daiichi Sankyo: Consultancy; Eisai: Consultancy; Janssen Pharmaceuticals: Consultancy; Astex Pharmaceuticals: Consultancy; Pfizer: Consultancy; Janssen Pharmaceuticals: Consultancy; Otsuka: Consultancy; Jazz Pharmaceuticals: Consultancy; Astex Pharmaceuticals: Consultancy; Cellectis: Research Funding; Roche/Genentech: Consultancy; Sandoz: Consultancy; Jazz Pharmaceuticals: Consultancy; Cellectis: Research Funding; Argenx: Consultancy; Aphivena Therapeutics: Consultancy. Sweet:Celgene: Honoraria, Speakers Bureau; BMS: Honoraria; Agios: Consultancy; Phizer: Consultancy; Agios: Consultancy; Celgene: Honoraria, Speakers Bureau; Astellas: Consultancy; Novartis: Consultancy, Honoraria, Speakers Bureau; Novartis: Consultancy, Honoraria, Speakers Bureau; Jazz: Speakers Bureau; BMS: Honoraria; Astellas: Consultancy; Phizer: Consultancy; Jazz: Speakers Bureau. Tallman:Cellerant: Research Funding; AROG: Research Funding; Orsenix: Other: Advisory board; AbbVie: Research Funding; ADC Therapeutics: Research Funding; BioSight: Other: Advisory board; Daiichi-Sankyo: Other: Advisory board.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal